library(tidyverse); library(patchwork)Environmental metadata
Import metadata & combine
tmp_all_metadata <- read.csv("input-data/All_metadata.csv")
# colnames(tmp_all_metadata)
# unique(tmp_all_metadata$Site)
# View(tmp_all_metadata)
july <- read.delim("input-data/flor_env_ALOHAJuly_SPOT.txt")
march <- read.delim("input-data/flour_ALOHAMarch.txt")# head(july)
# head(march)
fluor <- july %>%
left_join(march) %>%
pivot_longer(cols = SPOT:ALOHAMarch, names_to = "Site", values_to = "Fluor")Joining with `by = join_by(Depth)`# unique(fluor$Site)
metadata_all <- tmp_all_metadata %>%
filter((!Site == "ALOHA")) %>%
select(-X) %>%
left_join(fluor) %>%
pivot_longer(cols = Temp:Fluor, names_to = "VARIABLE", values_to = "VALUE")Joining with `by = join_by(Site, Depth, Fluor)`# head(metadata_all)
# unique(metadata_all$Site)Figure aesthetics
site_order <- c("SPOT", "PortofLA", "Catalina", "ALOHAJuly", "ALOHAMarch")
site_label <- c("SPOT", "Port of LA", "Catalina", "ALOHA July", "ALOHA March")
site_color_fill <- c("#3288bd", "#8073ac", "#66bd63", "#e6f598", "#d53e4f")
site_color_fill_nopola <- c("#3288bd", "#66bd63", "#e6f598", "#d53e4f")Isolate specific depths, add a label
spot<-c(1,150,885)
spotsite<-c("SPOT", "Catalina", "PortofLA")
aloha<-c(1,120,150,1000)
metadata_pt_depth <- metadata_all %>%
filter(Site %in% spotsite & Depth %in% spot | grepl("ALOHA", Site) & Depth %in% aloha) %>%
add_column(Label = "discrete") %>%
mutate(VAR_ORDER = factor(VARIABLE, levels = c("Temp", "Fluor", "Sal", "Oxy"), labels = c("Temperature", "Chlorophyll", "Salinity", "Oxygen"))) %>%
mutate(SITE_ORDER = factor(Site, levels = site_order, labels = site_label))
# unique(metadata_all$Site)allenv <- metadata_all %>%
mutate(VAR_ORDER = factor(VARIABLE, levels = c("Temp", "Fluor", "Sal", "Oxy"), labels = c("Temperature", "Chlorophyll", "Salinity", "Oxygen"))) %>%
mutate(SITE_ORDER = factor(Site, levels = site_order, labels = site_label)) %>%
arrange(Depth) %>%
ggplot(aes(y = Depth, x = VALUE)) +
geom_hline(yintercept = 0, alpha = 0.3) +
geom_path(size = 1, aes(color = SITE_ORDER)) +
geom_point(data = metadata_pt_depth, size = 2, aes(y = Depth, x = VALUE, fill = SITE_ORDER, shape = SITE_ORDER)) +
scale_shape_manual(values = c(21, 22, 23, 24, 25)) +
scale_fill_manual(values = site_color_fill) +
scale_color_manual(values = site_color_fill) +
scale_y_reverse() +
facet_grid(. ~ VAR_ORDER, scales = "free", switch = "both") +
theme_linedraw() +
labs(x = "", y = "Depth (m)") +
theme(strip.background = element_blank(),
strip.placement = "outside",
strip.text = element_text(color = "black"),
legend.title = element_blank())Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.allenvWarning: Removed 2 rows containing missing values (`geom_path()`).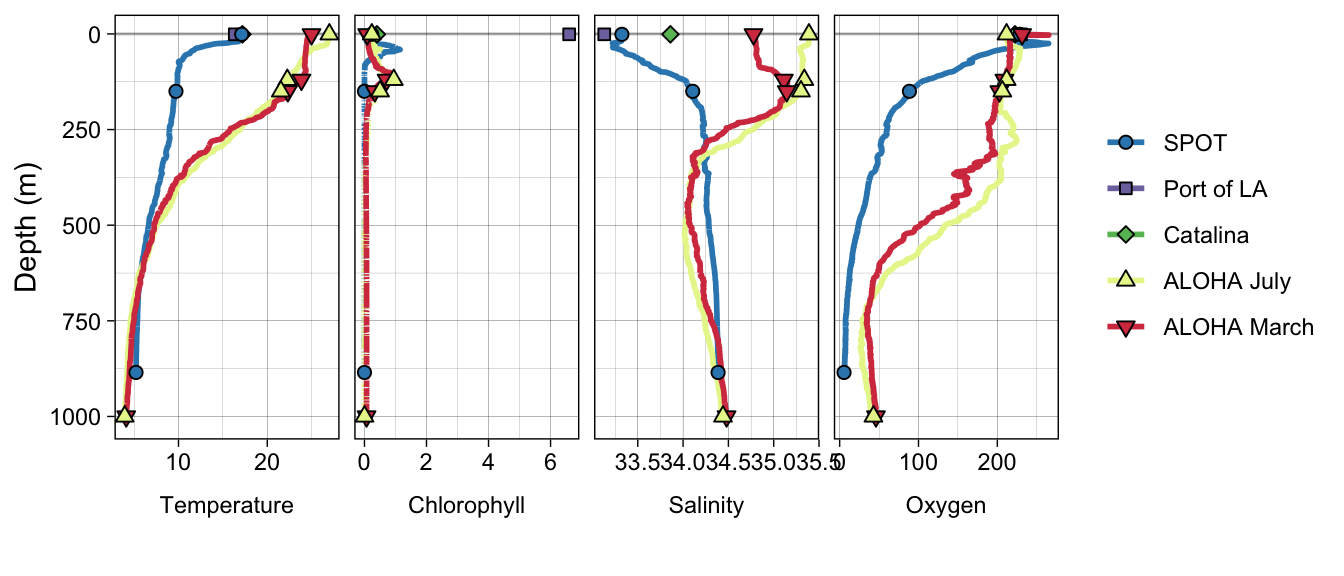
no_pola <- metadata_all %>%
mutate(VAR_ORDER = factor(VARIABLE, levels = c("Temp", "Fluor", "Sal", "Oxy"), labels = c("Temperature", "Chlorophyll", "Salinity", "Oxygen"))) %>%
mutate(SITE_ORDER = factor(Site, levels = site_order, labels = site_label)) %>%
arrange(Depth) %>%
filter(Site != "PortofLA") %>%
ggplot(aes(y = Depth, x = VALUE)) +
geom_hline(yintercept = 0, alpha = 0.3) +
geom_path(size = 1, aes(color = SITE_ORDER)) +
geom_point(data = (metadata_pt_depth %>% filter(Site != "PortofLA")), size = 2, aes(y = Depth, x = VALUE, fill = SITE_ORDER, shape = SITE_ORDER)) +
scale_shape_manual(values = c(21, 22, 23, 24, 25)) +
scale_fill_manual(values = site_color_fill_nopola) +
scale_color_manual(values = site_color_fill_nopola) +
scale_y_reverse() +
facet_grid(. ~ VAR_ORDER, scales = "free", switch = "both") +
theme_linedraw() +
labs(x = "", y = "Depth (m)") +
theme(strip.background = element_blank(),
strip.placement = "outside",
strip.text = element_text(color = "black"),
legend.title = element_blank())
no_polaWarning: Removed 2 rows containing missing values (`geom_path()`).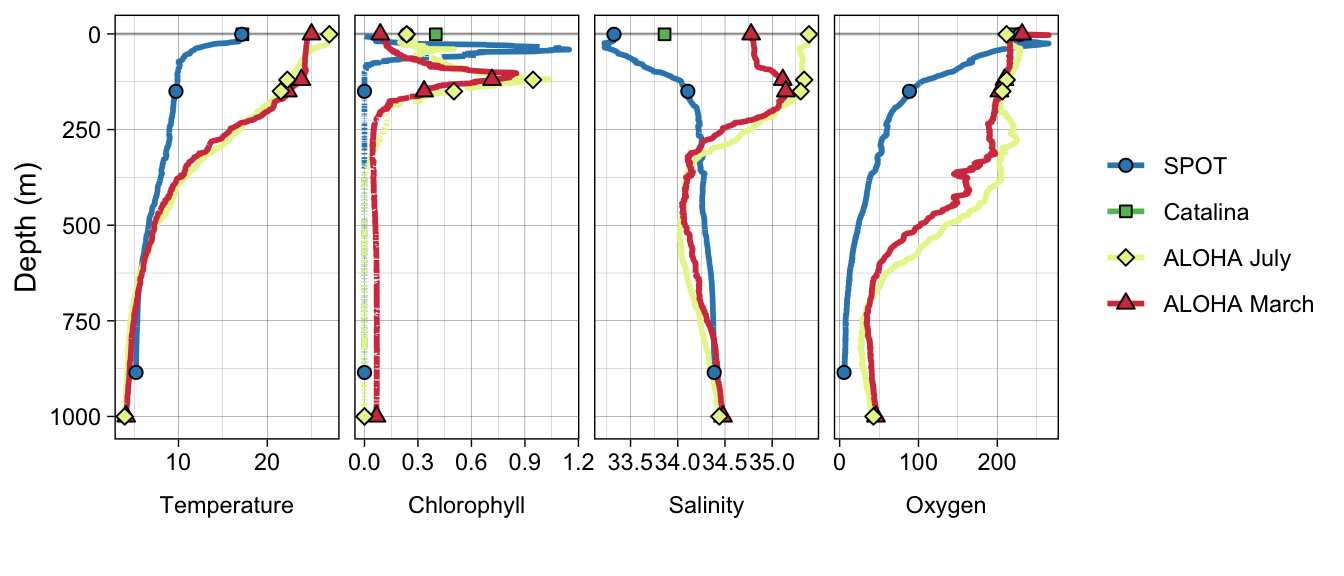
# svg("figs/water-col.svg", w = 8, h = 7)
allenv + no_pola + patchwork::plot_layout(ncol = 1)Warning: Removed 2 rows containing missing values (`geom_path()`).
Removed 2 rows containing missing values (`geom_path()`).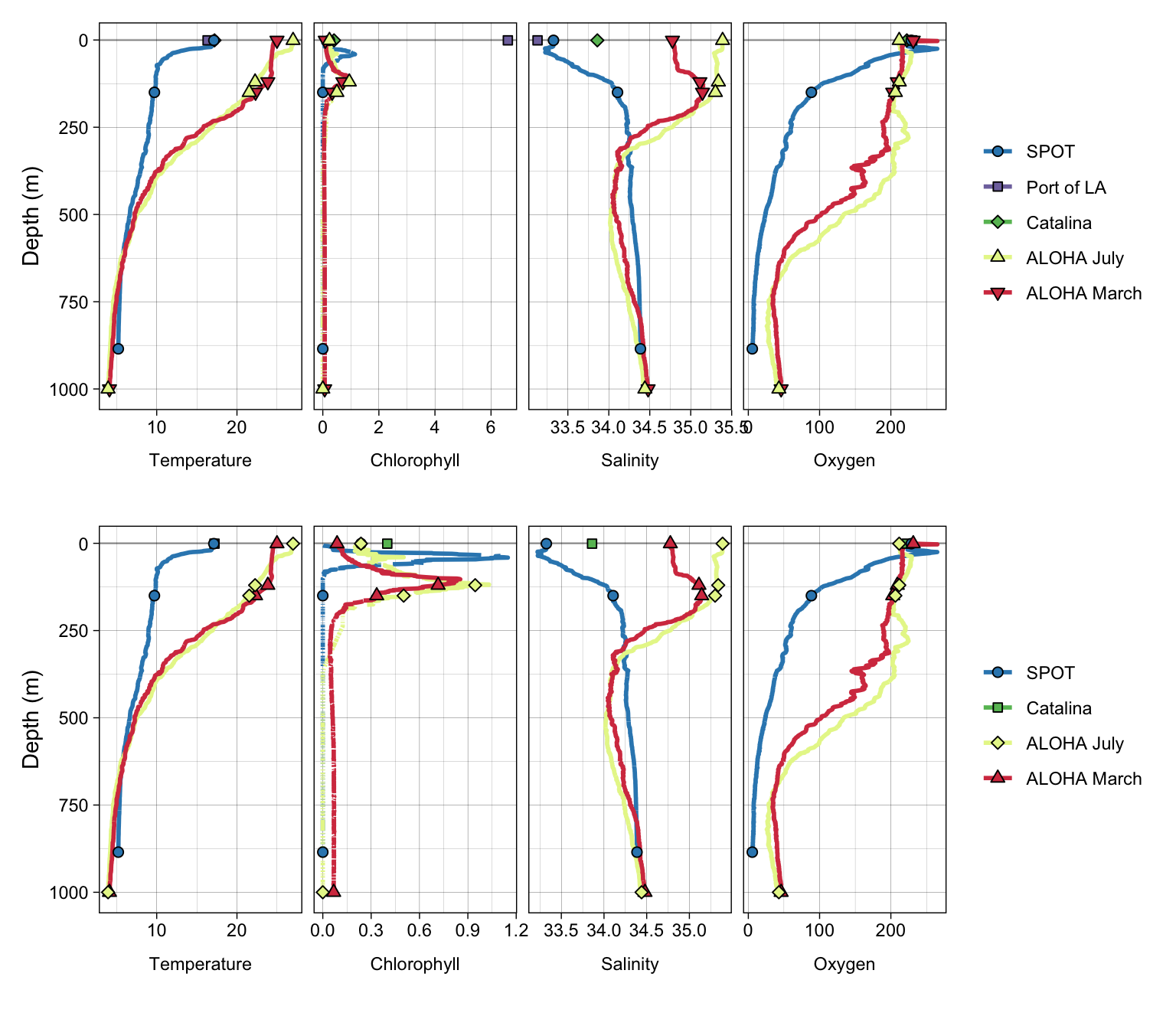
# dev.off()
# Requires manual modification of chlorophyll with Port of LA sample